It’s 2 AM in a lab somewhere. A researcher has three terminals open, a half-written Jupyter notebook on one screen, an Excel sheet filled with sample IDs on another, and a half-eaten snack next to shell commands. They’re juggling scripts to run a protein folding model, parsing CSVs from the last experiment, searching for literature, and Googling whether that one Python package broke in the latest update, again.
This isn’t the exception; it’s the norm. Scientific research today is a patchwork of tools, scripts, and formats, glued together by determination and late-night caffeine. Reproducibility is a wishlist item. Infrastructure is an afterthought. And while automation exists, it’s usually hand-rolled and stuck on someone’s laptop.
But what if science workflows could be orchestrated, end-to-end, by an intelligent agent?
What if instead of writing shell scripts and hoping the dependencies don’t break, a scientist could describe the goal, “read this CSV of compounds and proteins, search for literature, admet, and more”, and an AI agent could plan the steps, spin up the right tools in containers, execute the tasks, and even summarize the results?
That’s the promise of science agents. AI-powered systems that don’t just answer questions like ChatGPT, but autonomously carry out entire research workflows. And thanks to the convergence of LLMs, GPUs, Dockerized environments, and open scientific tools, this shift isn’t theoretical anymore.
It’s happening now.

What is a Science Agent?
A Science Agent is more than just a chatbot or a smart prompt generator; it’s an autonomous system designed to plan, execute, and iterate on entire scientific workflows with minimal human input.
Instead of relying on one-off questions like “What is ADMET?” or “Summarize this paper,” a science agent operates like a digital research assistant. It understands goals, breaks them into steps, selects the right tools, runs computations, and even reflects on results.
CrewAI: AI agents framework -> https://www.crewai.com/
ADMET: how a drug is absorbed, distributed, metabolized, and excreted, and its toxicity
Let’s make it concrete:
Take this multi-agent system you might build with CrewAI:
- Curator: Data-focused agent whose primary role is to ensure data quality and standardization.
- Researcher: Literature specialist. Its main goal is to find relevant academic papers on PubMed for the normalized entities provided by the Curator.
- Web Scraper: Specialized agent for extracting information from websites.
- Analyst: Predicts ADMET properties and toxicity using models or APIs.
- Reporter: Compiles all results into a clean Markdown report.
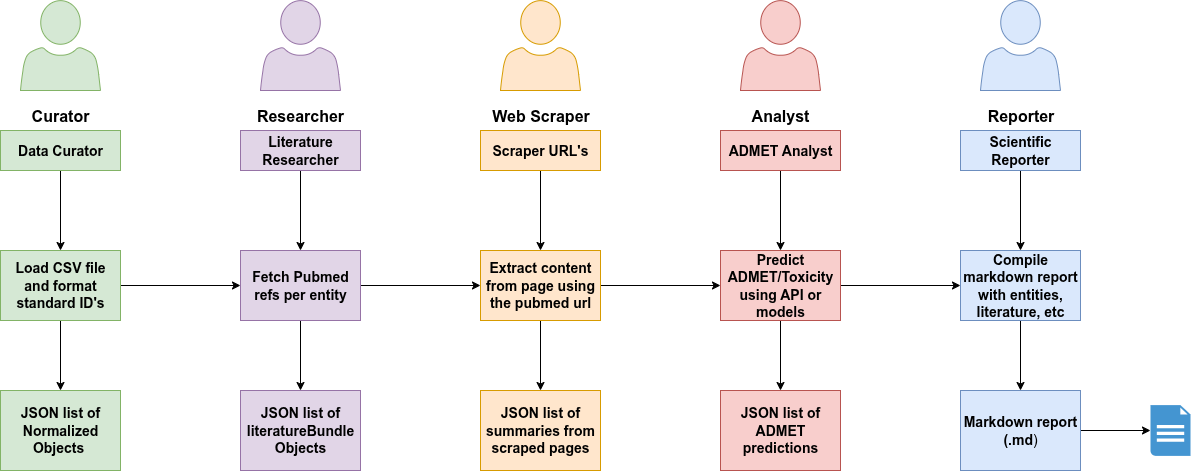
Each of these agents acts independently but works as part of a coordinated system. Together, they automate what would take a human team hours or even days, now in minutes and reproducibly.
Why This Is Different from ChatGPT
You’ve probably used ChatGPT to summarize papers, write Python code, or explain complex topics. And while it might seem like a simple question-answer engine, there’s often more happening behind the scenes, prompt chains, context windows, and latent loops of reasoning. But even with those advances, these interactions are still mostly human-in-the-loop: you ask, it answers.
Science agents are a different species entirely.
Instead of waiting for your next prompt, they plan and execute entire workflows autonomously. They decide which tools to use based on context, how to validate results, and when to pivot. Where ChatGPT responds, agents act. They’re less like assistants and more like collaborators.
Let’s break down the key differences:
|
Feature |
LLMs (ChatGPT & similar) |
Science Agents (CrewAI, LangGraph, etc.) |
|---|---|---|
|
Interaction |
Multi-turn, often guided by user prompts or system instructions |
Long-running, autonomous workflows across multiple tools |
|
Role |
Assistant with agentic capabilities abstracted away |
Explicit research collaborator executing role-specific tasks |
|
Autonomy |
Semi-autonomous; requires external prompting or embedded system orchestration |
Fully autonomous planning, tool selection, and iteration |
|
Tool Use |
Some tools are used via plugins/functions (e.g., browser, code interpreter) |
Explicit tool integration (APIs, simulations, databases, Dockerized tools) |
|
Memory |
Short- to medium-term context (limited per session or chat, non-explicit workspace) |
Persistent long-term memory (vector DBs, file logs, databases, explicit and programmable) |
|
Reproducibility |
Very limited, without the ability to define agents’ roles/tasks and their tools |
Fully containerized, versioned workflows, reproducible workflows with defined agent roles/tasks |
Try it yourself
If you’re curious, here’s a two-container demo you can run in minutes.
git repo: https://github.com/estebanx64/docker_blog_ai_agents_research
We just have two containers/services for this example:
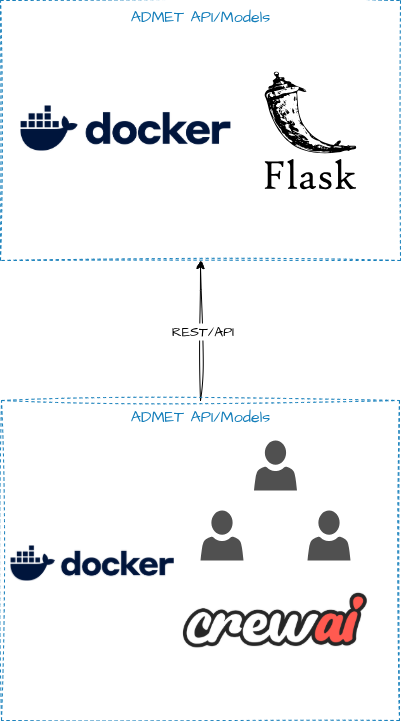
Prerequisites
- Docker and Docker Compose
- OpenAI API key (for GPT-4o model access)
- Sample CSV file with biological entities
Follow the instructions from README.md in our repo to set up your OpenAI API KEY
Running the next workflow with the example included in our repo is going to charge ~1-2 USD for the OpenAI API.
Run the workflow.
docker compose up
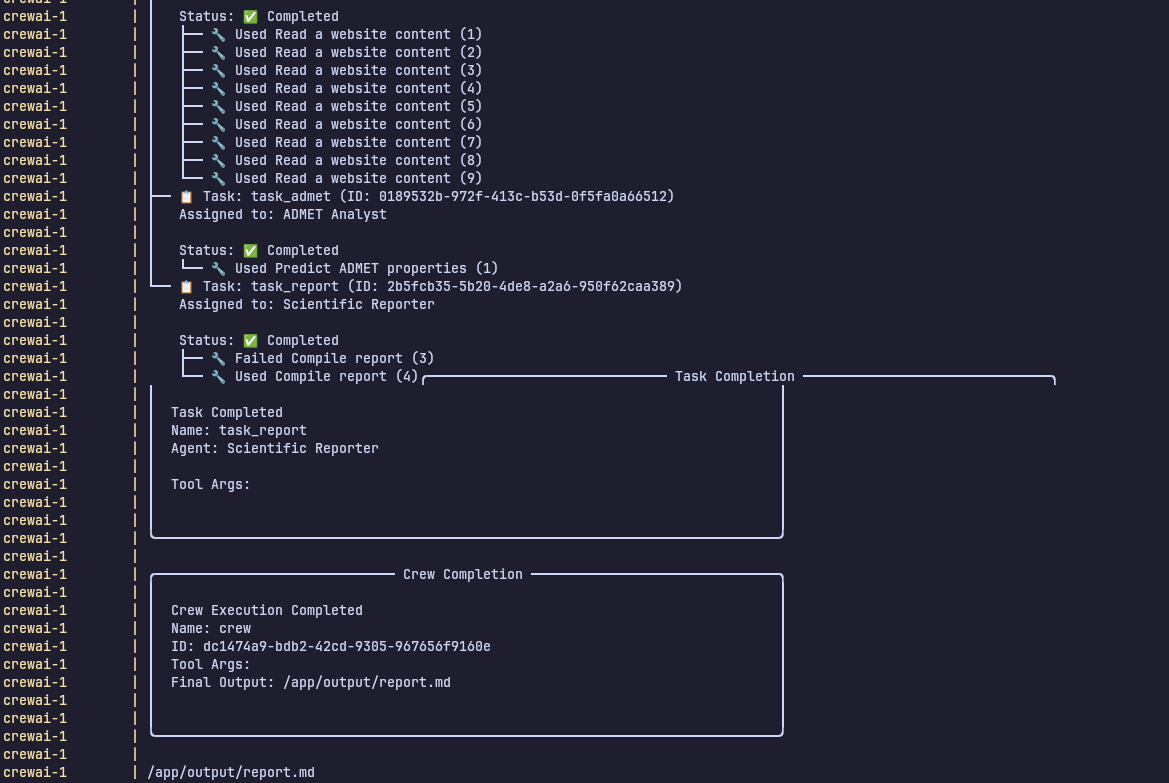
The logs above demonstrate how our agents autonomously plan and execute a complete workflow.
Ingest CSV File
- The agents load and parse the input CSV dataset.
Query PubMed
- They automatically search PubMed for relevant scientific articles.
Generate Literature Summaries
- The retrieved articles are summarized into concise, structured insights.
Calculate ADMET Properties
- The agents call an external API to compute ADMET (Absorption, Distribution, Metabolism, Excretion, Toxicity) predictions.
Compile Results into Markdown Report
- All findings are aggregated and formatted into a structured
report.md.
Output Files
report.md– Comprehensive research report.- JSON files – Contain normalized entities, literature references, and ADMET predictions.
This showcases the agents’ ability to make decisions, use tools, and coordinate tasks without manual intervention.
If you want to explore and dive in more, please check the README.md included in the github repository
Imagine if your lab could run 100 experiments overnight, what would you discover first?
But to make this vision real, the hard part isn’t just the agents, it’s the infrastructure they need to run.
Infrastructure: The Bottleneck
AI science agents are powerful, but without the right infrastructure, they break quickly or can’t scale. Real research workflows involve GPUs, complex dependencies, and large datasets. Here’s where things get challenging, and where Docker becomes essential.
The Pain Points
- Heavy workloads: Running tools like AlphaFold or Boltz requires high-performance GPUs and smart scheduling (e.g., EKS, Slurm).
- Reproducibility chaos: Different systems = broken environments. Scientists spend hours debugging libraries instead of doing science.
- Toolchain complexity: Agents rely on multiple scientific tools (RDKit, PyMOL, Rosetta, etc.), each with their own dependencies.
- Versioning hell: Keeping track of dataset/model versions across runs is non-trivial, especially when collaborating.
Why Containers Matter
- Standardized environments: Package your tools once, run them anywhere, from a laptop to the cloud.
- Reproducible workflows: Every step of your agent’s process is containerized, making it easy to rerun or share experiments.
- Composable agents: Treat each step (e.g., literature search, folding, ADMET prediction) as a containerized service.
- Smooth orchestration: You can use the CrewAI or other frameworks’ capabilities to spin up containers and isolate tasks that need to run or validated output code without compromising the host.
Open Challenges & Opportunities
Science agents are powerful, but still early. There’s a growing list of challenges where developers, researchers, and hackers can make a huge impact.
Unsolved Pain Points
- Long-term memory: Forgetful agents aren’t useful. We need better semantic memory systems (e.g., vector stores, file logs) for scientific reasoning over time.
- Orchestration frameworks: Complex workflows require robust pipelines. Temporal, Kestra, Prefect, and friends could be game changers for bio.
- Safety & bounded autonomy: How do we keep agents focused and avoid “hallucinated science”? Guardrails are still missing.
- Benchmarking agents: There’s no standard to compare science agents. We need tasks, datasets, and metrics to measure real-world utility.
Ways to Contribute
- Containerize more tools (models, pipelines, APIs) to plug into agent systems.
- Create tests and benchmarks for evaluating agent performance in scientific domains.
Conclusion
We’re standing at the edge of a new scientific paradigm, one where research isn’t just accelerated by AI, but partnered with it. Science agents are transforming what used to be days of fragmented work into orchestrated workflows that run autonomously, reproducibly, and at scale.
This shift from messy shell scripts and notebooks to containerized, intelligent agents isn’t just about convenience. It’s about opening up research to more people, compressing discovery cycles, and building infrastructure that’s as powerful as the models it runs.
Science is no longer confined to the lab. It’s being automated in containers, scheduled on GPUs, and shipped by developers like you.
Check out the repo and try building your own science agent. What workflow would you automate first?







