This is a guest blog post from Dr. Kelsey Florek and Curtis Kapsak. A video presentation of this post from Docker’s Community All-Hands can be found here.
Dr. Kelsey Florek is the Senior Genomics and Data Scientist at the Wisconsin State Laboratory of Hygiene and a steering committee member of the State Public Health Bioinformatics group (StaPH-B).
Curtis Kapsak is a Bioinformatics Scientist at Theiagen Genomics and is a member of the State Public Health Bioinformatics group (StaPH-B).
The rapid appearance and global spread of a novel Severe Acute Respiratory Syndrome (SARS) virus in 2019 pushed public health laboratories to develop new methods for genomic monitoring efforts on a scale never seen before. Adding to this challenge, the approaches typically used in genomic data analysis often rely on cutting edge and often niche open source software and libraries that increase the complexity of setting up analytical pipelines or workflows. This along with a varying landscape of compute environments ranging from on-prem workstations to public Cloud created a significant barrier for many laboratories attempting to perform viral genomic monitoring.
Developing a library of dockerized tools
Public health laboratories inherently need to meet rigorous quality control and quality assurance standards. The tests performed in public health laboratories are either reported back to clinics to be used for patient care or used in aggregate to inform public health interventions or outbreak investigations. Analytical workflows are held to the same standards as other laboratory developed tests and to support this effort, the State Public Health Bioinformatics consortium (StaPH-B) started developing a repository of dockerized software that was commonly used in public health genomic data analyses, StaPH-B/docker-builds. This repository was designed to address the need for accessible software that is both highly reliable and reproducible. Combined with a usage guide, this repository provided a centralized location of maintained and tested open source tools to support laboratories developing analysis workflows.
Since its initial development in 2018 the StaPH-B/docker-builds repository has grown to contain multiple versions of over 90 different analytical tools from 19 different contributors, with several of the COVID-19 specific images achieving over 1 million pulls. Between March 2021 and January 2022 as more laboratories began genomic monitoring we saw a logarithmic increase in the number of Docker image pulls on core COVID-19 genomic analysis software.
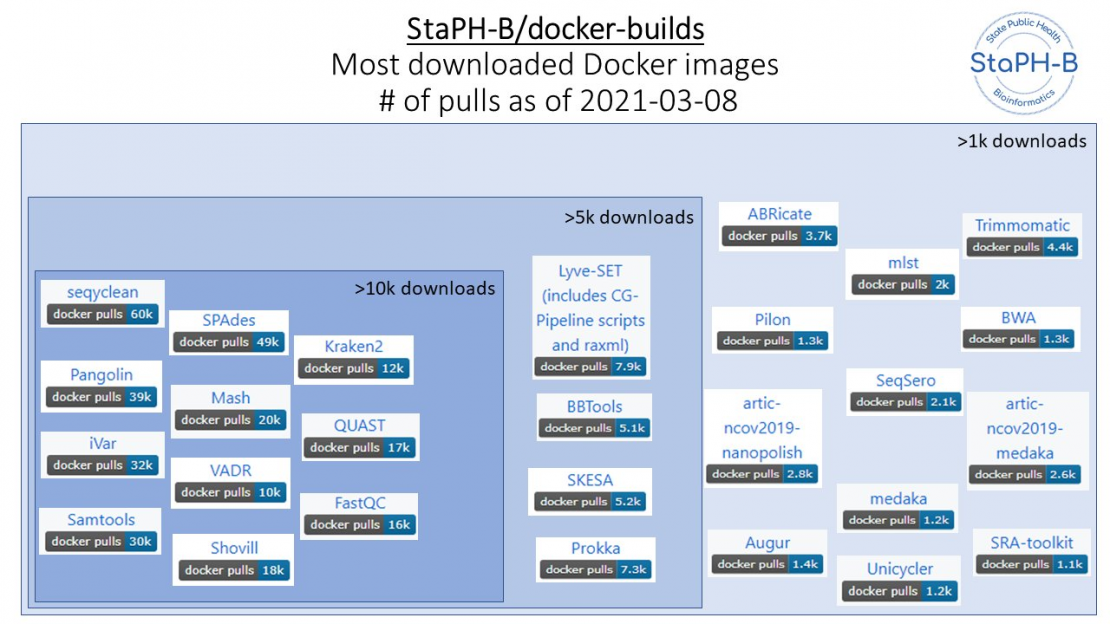
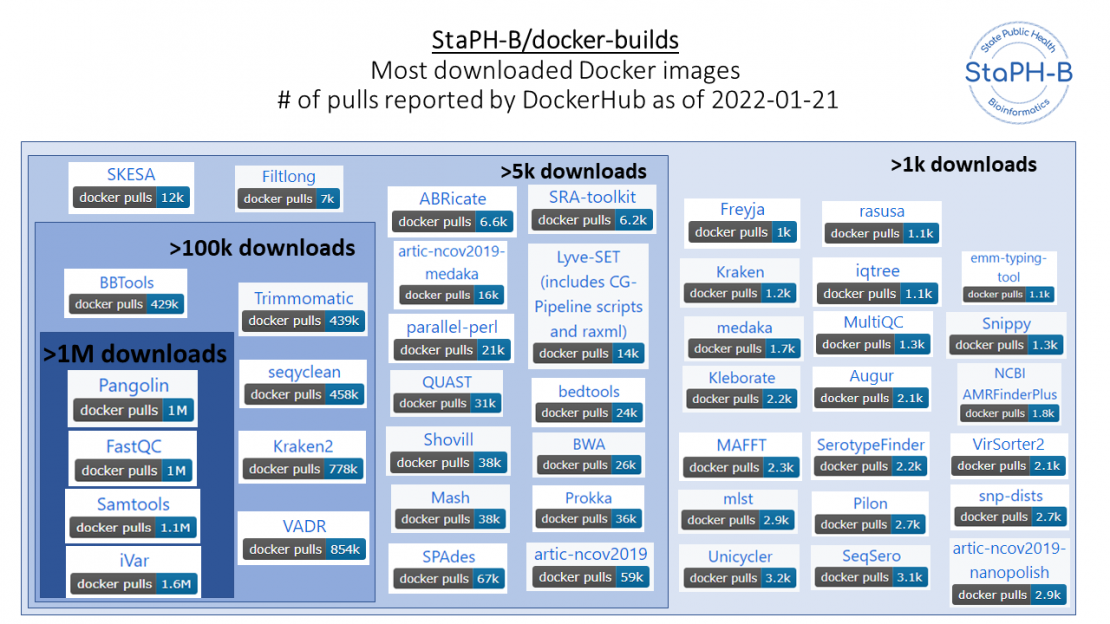
Supporting COVID-19 genomic analytics
Bioinformatic pipelines or workflows consist of a variety of tools and often start from a form of raw or primary DNA sequencing data. These tools perform a variety of transformative or summary tasks and vary in both their computational requirements and dependencies. The process of sequencing the SARS-CoV-2 viral genome involves sectioning off the viral genome and sequencing small portions of the DNA in parallel. The result is a dataset containing hundreds of thousands to millions of short strings containing A’s, T’s, C’s, and G’s in a variety of sequence combinations. COVID-19 workflows then take these datasets, reconstruct the genome and use a variety of techniques to then characterize the virus.
Many laboratories across the globe have moved towards using a dedicated workflow language like WDL or Nextflow for their analytical workflows. Combining a workflow language with dockerized software allows for the creation and routine usage of workflows that are highly portable and easily adapted to a variety of compute environments. This gives laboratories the ability to run small datasets on a laptop or scale to a high performance compute cluster or cloud environment for large datasets. Additionally, these workflow approaches allow developing a modular analysis framework that enables swapping out software as new versions are released or issues are identified. With the rapid and constant evolution of the virus that causes COVID-19, updates to classification software are also frequently updating to maintain the ability to accurately identify variants.
Keeping up to speed with viral evolution
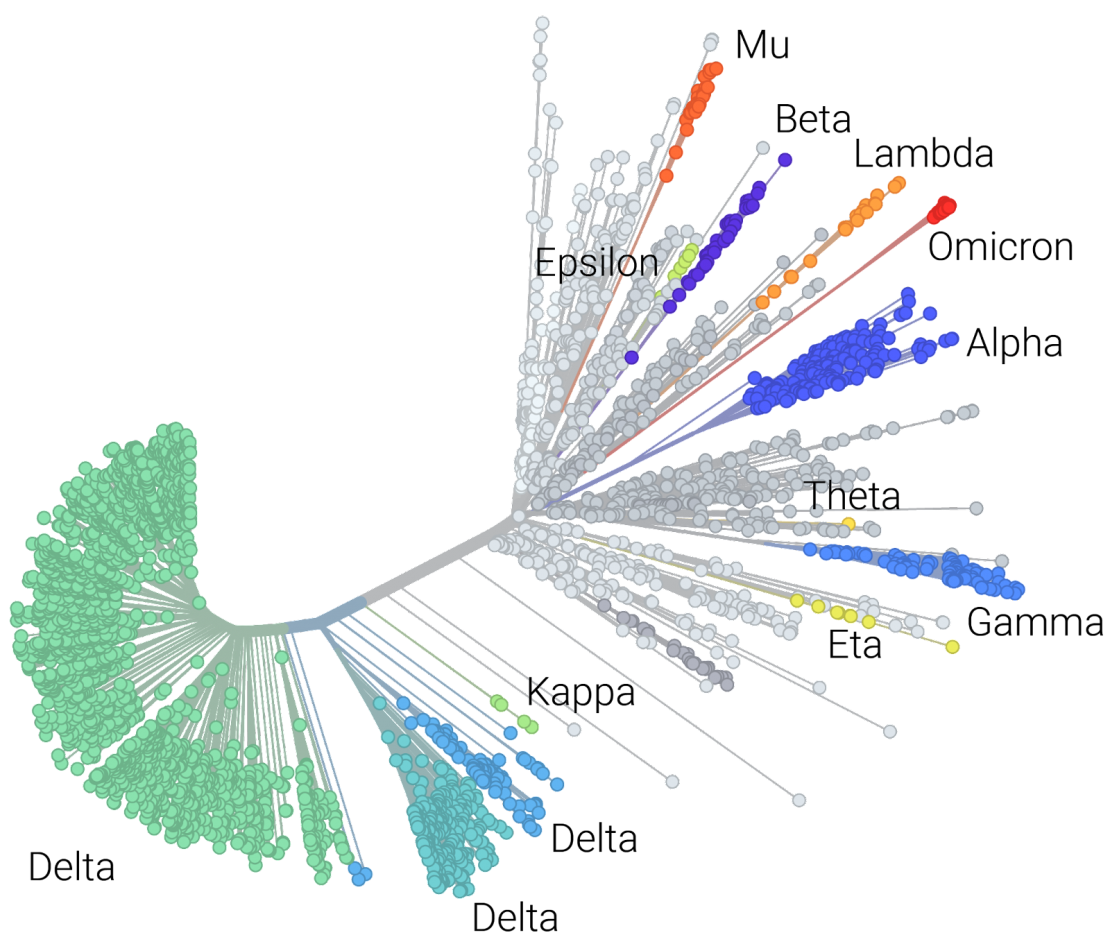
The COVID-19 virus evolves a bit slower than influenza accruing on average two mutations per month and different variants (Alpha, Delta, Omicron, etc.) are differentiated by various combinations of mutations. Classifying a virus requires constructing a phylogenetic tree that models the relationship of the new virus to other viruses. However, constructing a tree to compare each new virus to every previous virus is both computationally expensive and impractical. To address this, two commonly used methods have emerged including using a set of selected reference viruses to build a tree (Nextclade) or machine learning to classify mutational patterns (Pangolin). Both of these approaches require regular updates to ensure classification is occurring accurately with the most recent information. Leveraging containerization, StaPH-B has been able to maintain images with the latest models allowing users to run workflows knowing they are using the most up to date, robust, and tested classification tools.
Summary
The highly portable, scaleable, and efficient nature of containerization has transformed how public health disease monitoring is performed. The implementation of containerized workflows have enabled laboratories to quickly adopt complex analytical workflows, which in turn has grown the scale of the viral monitoring effort. The open source repository maintained by StaPH-B would not be possible without the community of bioinformaticians driving innovation. With more laboratories turning to sequencing and complex analytics there is a growing demand for people to bridge the gap between biology and informatics. If you are interested in a career in bioinformatics and using data to solve health problems be sure to check out the APHL-CDC Bioinformatics Fellowship!
DockerCon2022
Join us for DockerCon2022 on Tuesday, May 10. DockerCon is a free, one day virtual event that is a unique experience for developers and development teams who are building the next generation of modern applications. If you want to learn about how to go from code to cloud fast and how to solve your development challenges, DockerCon 2022 offers engaging live content to help you build, share and run your applications. Register today at https://www.docker.com/dockercon/
